SOLA Enzymatic DNA Synthesis Technology
The challenge in oligonucleotide synthesis
Researchers need DNA oligonucleotides for a wide variety of life science applications but obtaining them is fraught with challenges. Most scientists order oligos from vendors, but that can be costly and slow. Oligos have historically been built by just a few centralized manufacturers, and their processes are very capital intensive and typically require outdated methods using toxic chemicals.
A new approach is enzymatic DNA synthesis (EDS), which is safer and therefore amenable for de-centralized oligo production at the benchtop. Telesis Bio is building on its history of innovation to develop a new generation of enzymatic DNA synthesis known as SOLA (short oligonucleotide ligation assembly), which will be integrated into our next generation Oligo Printer and Digital to Biological Converter instruments. Due to the high-fidelity, low-cost nature of the approach, SOLA is the only EDS platform that is scalable into synthetic biology applications.
Supercharge discovery with on-demand oligo production
SOLA technology enables rapid turnaround time for high-quality, low-cost synthetic DNA ranging from oligonucleotides up to complete genomes. Combined with the BioXp™ 3250 system, SOLA is the next step on the path to making desktop printing of biology a reality.
The SOLA method begins with a digital DNA sequence to perform rapid, high-fidelity assembly of DNA oligos. The scalable technique produces oligos up to 100 bp, allowing for the on-demand production of PCR primers, diagnostic probes, sequencing enrichment probes and CRISPR-Cas9 guide RNAs. In addition, overlapping 100 bp products can be rapidly assembled on the BioXp™ system to produce synthetic genes, genomes, mRNA and protein enabling fully-automated digital to biological conversion on your benchtop.
On-demand full-length gene, mRNA and protein synthesis
BioXp DBC Systems* will contain all the reagents necessary to go from digital gene sequence to the production of biologically active DNA, mRNA and protein at unprecedented speed.
How SOLA Enzymatic DNA Synthesis (EDS) Process Works
SOLA uses a universal, pre-manufactured library of short DNA building blocks to construct oligo sequences up to 100 bp in length. For applications requiring longer builds such as gene, mRNA, or protein synthesis the 100-bp products can be further processed using well-established methods such as Gibson Assembly®.
Beginning with a standard collection of only a few thousand sequences with variable ends, any DNA sequence can be made. Depending on the application, the building blocks reside in high-density microtiter plates. Upon entering the desired DNA sequence, software determines which building blocks are required.
This process involves just three cycles of a two-step process that includes PCR amplification and a concerted digestion/ligation reaction. This cyclical process is repeated up to three more times to form a 100-base sequence. In contrast, traditional approaches that use individual bases would require 100 cycles of a 4-step process.
SOLA EDS enables robust and reproducible oligo synthesis
SOLA uses a universal, pre-manufactured library of short DNA building blocks to construct oligo sequences up to 100 bp in length. For applications requiring longer builds such as gene, mRNA, or protein synthesis the 100-bp products can be further processed using well-established methods such as Gibson Assembly.
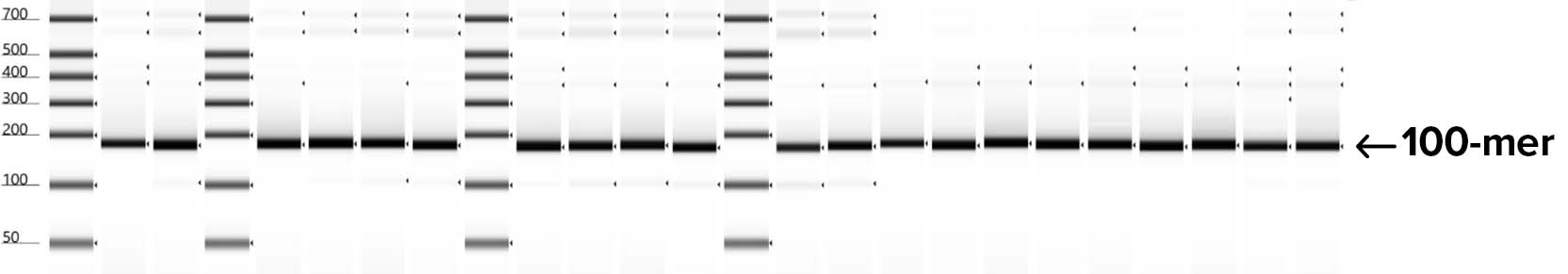
Automated electrophoresis of oligos to check for quality and size of fragments synthesized shows 100% success rate using SOLA EDS for building 24X 100-mer replicates for a segment of the HA flu gene. The products shown on this gel represent 100 bp of synthetic DNA nested between flanking regions that can be removed by restriction digestion prior to gene assembly
Advantages over alternative enzymatic DNA synthesis chemistries
Conventional oligo synthesis approaches that use traditional phosphoramidite chemistry or terminal deoxynucleotidyl transferase (TdT) incorporate a single DNA base at a time. Because SOLA stitches together purified and sequence-verified DNA blocks to synthesize oligos, it’s significantly faster, more accurate, and higher-throughput than its predecessors. In addition, because these DNA blocks can be manufactured in large quantities and used hundreds of thousands of times before replenishing, the DNA synthesis costs are driven down exponentially.
SOLA also offers these benefits without the hazardous waste associated with the phosphoramidite chemistry used in traditional oligo synthesis pipelines. Therefore, SOLA has the potential to be the best enabling technology for the on-demand desktop printing of biology across life sciences and synthetic biology, while also making a positive environmental impact.
Fidelity
Coupling efficiencies approaching 100% result in 50x improvement in fidelity compared to alternative methods. This means scientists can build DNA, mRNA and proteins accurately.
Build length
Can construct short DNA fragments for CRISPR guide RNAs up to full-length gene fragments. Fragments generated with SOLA can also be used for gene pathways, whole genomes and templates for mRNA and protein synthesis.
Flexibility
Flexible workflow allows the process to be stopped at various size increments. Libraries for specific applications enable even more possibilities spanning life sciences and synthetic biology applications.
Throughput
Rapid & robust workflow enables higher throughput than traditional DNA synthesizers. This enables even more design, build & test cycles than ever before.
Cost
Workflow is based on standard off-the-shelf enzymes and high-yield, low-cost DNA libraries. This means lower per base cost for DNA synthesis applications.
Consistently high coupling efficiencies enable high-fidelity oligo building blocks.
Higher fidelity oligos support high-fidelity gene, mRNA and protein synthesis
Combination of high coupling efficiencies and fidelity enable a broad array of synthetic biology applications
| Oligo length | CRISPR guides | PCR primers | NGS probes | Gene synthesis | mRNA synthesis | Protein synthesis |
|---|---|---|---|---|---|---|
| 10-mer | • | |||||
| 16-mer | • | |||||
| 28-mer | • | • | ||||
| 52-mer | • | |||||
| 100-mer | • | • | • | • | • |
SOLA offers robust performance for a variety of key applications
SOLA chemistry uses short DNA building blocks to synthesize oligos up to 100 bp in length, requiring only 3 cycles to complete. This highly differentiated technology offers significantly higher coupling efficiencies compared with other synthesis chemistries. This means oligos produced have greater sequence accuracy and higher percentage of full-length product which is critical for the accuracy of downstream oligo-dependent applications like CRISPR-based gene editing, target enrichment for next-generation sequencing, traditional primer-based PCR methods, and synthetic biology methods. This higher performance in oligo synthesis technology has the potential to positively impact the quality of a wide variety of molecular biology-based applications in the life sciences.
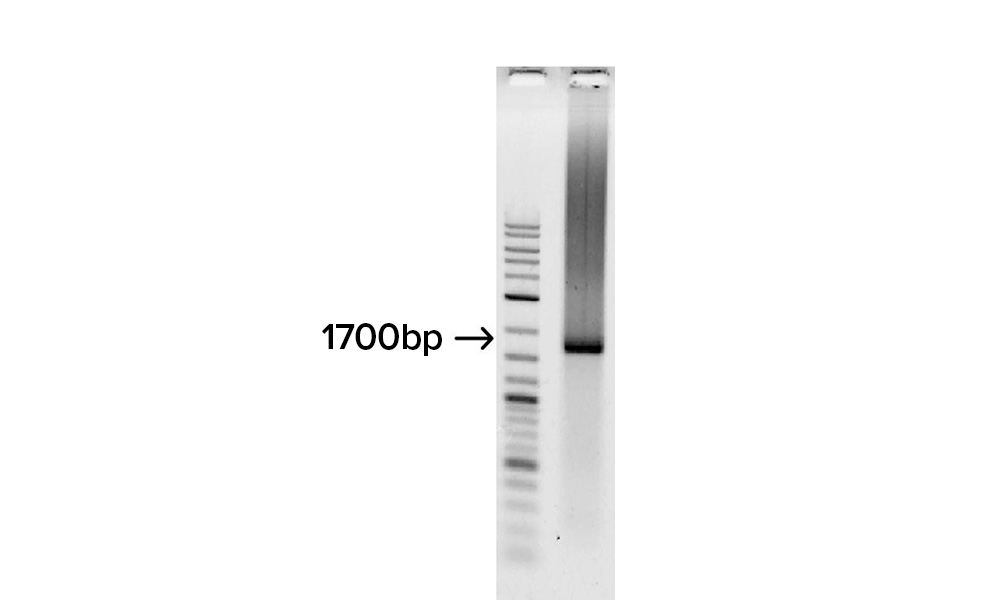
Gel image shows the assembly of the fully synthetic 1.7 kb H1 flu gene derived from 32 overlapping SOLA generated 100-mers.
The full-length gene was confirmed by cloning and DNA sequencing and determined to have an error rate of approximately 1 error per 6,000 bp, prior to applying enzymatic error correction.
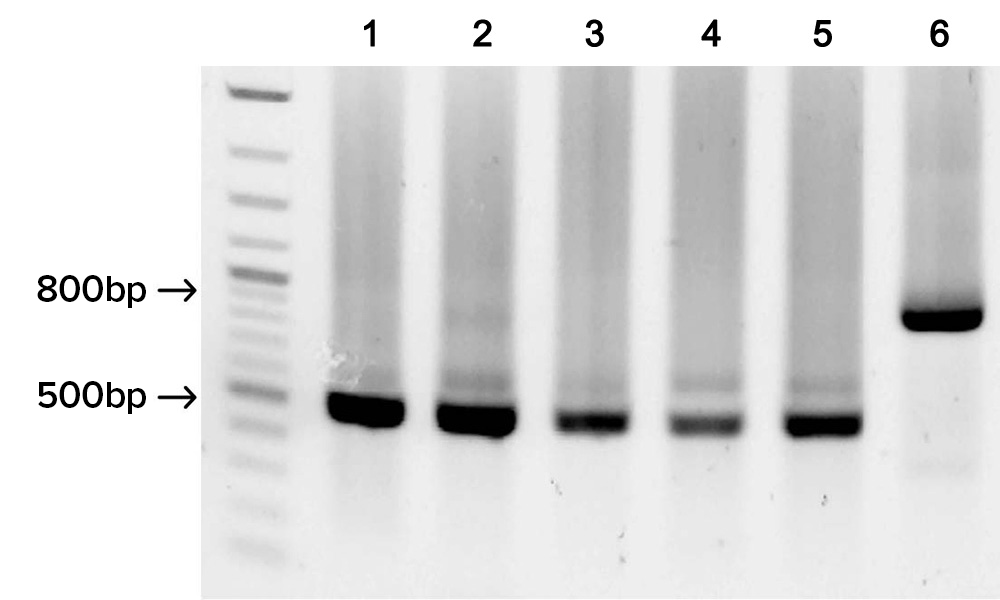
Gel image shows the successful assembly of five different fully-synthetic 500 bp HA flu gene fragments each derived from 9 overlapping SOLA generated 100-mers, and the assembly of one 800 bp HA flu gene product derived from 16 overlapping SOLA generated 100-mers.
Full length synthetic gene fragments were assembled and cloned using Gibson Assembly. DNA sequencing was used to determine an error rate of fewer than 1 error per 4,000 bp prior to applying enzymatic error correction.
A
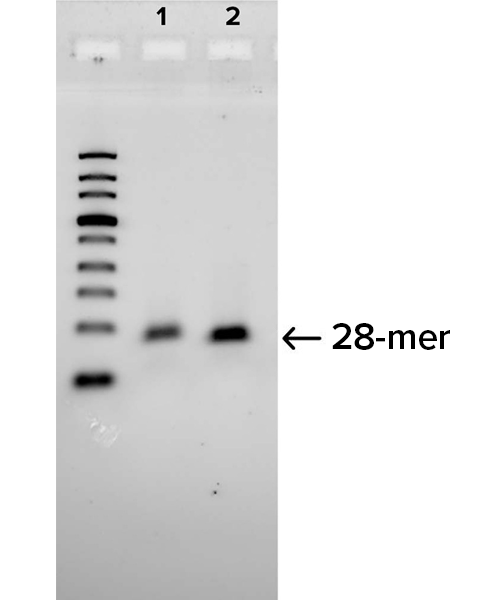
B
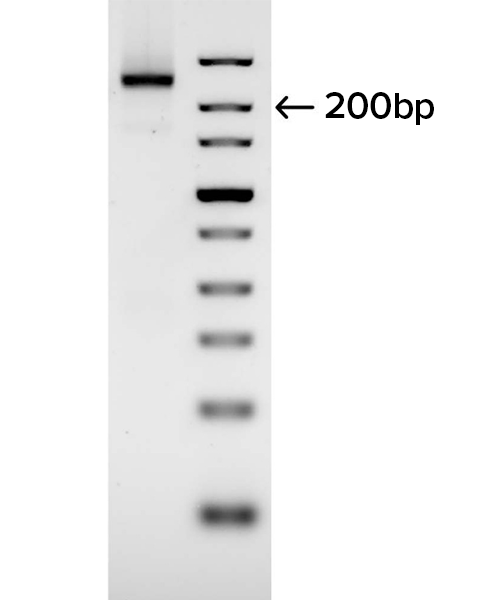
Gel images demonstrate the successful production of two 28-mer ssDNA PCR primers (A) and amplification of a 250 bp region of an E. coli gene (B) using the PCR primers shown in (A). Following two rounds of SOLA EDS to produce dsDNA 28-mers, the products were enzymatically converted to ssDNA and flanking regions were removed. The 250bp PCR product was confirmed by DNA sequencing with 100% of the colonies demonstrated to be error-free.
A
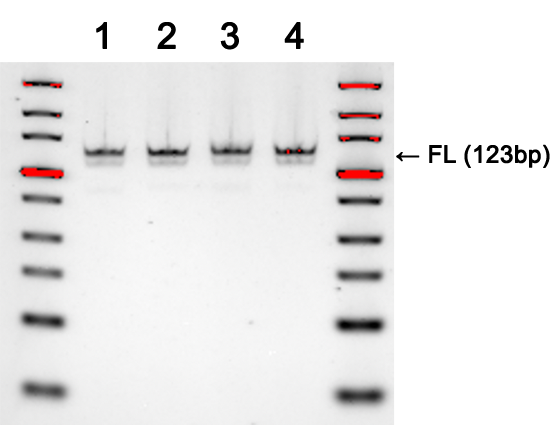
B
Traditional workflow
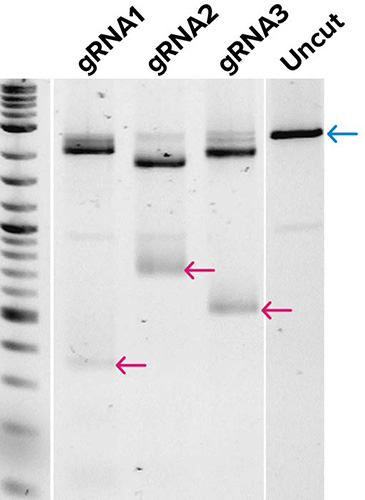
← Full-length product
SOLA EDS workflow
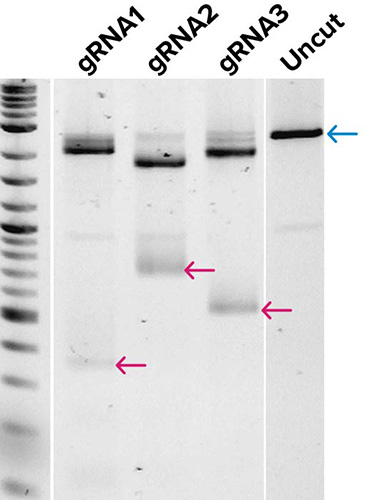
← Cas9 cleavage product (low mw)
Gel images demonstrate the production of DNA templates for four 20-mer guide RNAs (A), and the successful demonstration of biological activity of three guide RNAs generated from (A) as tested by in vitro digestion of a pUC19 plasmid DNA template when combined with Cas9 enzyme (B). Expected low mw products from Cas9 cleavage are indicated along with the uncut control, which shows the expected full-length (FL) product. Very little qualitative differences were observed in gRNAs from templates generated by traditional workflows or by SOLA EDS.